Interested in a paper but cant get it through your library?
Drop us an email and we will send you a pdf !
2024
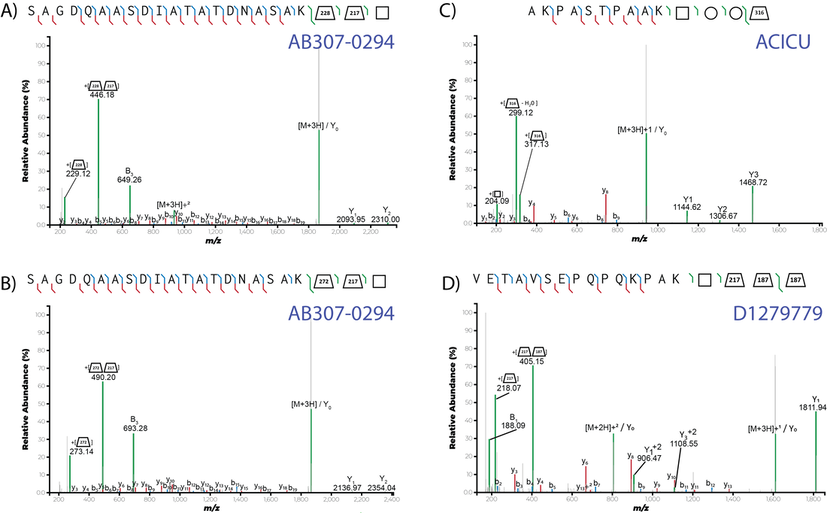
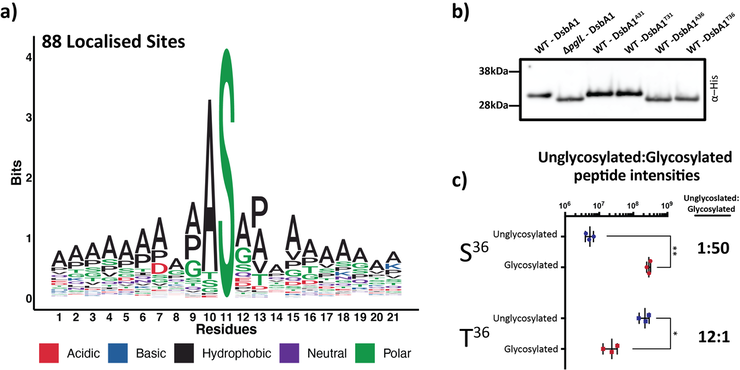
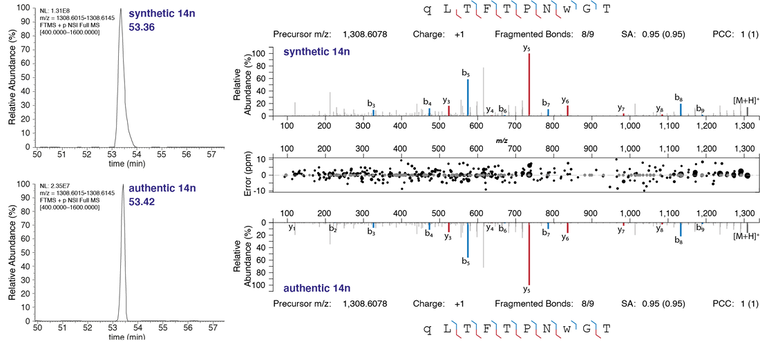
1. K. I. Tkalec, A. J. Hayes, K. S. Lim, J.M. Lewis, M. R. Davies, N. E. Scott* Glycan-tailored glycoproteomic analysis reveals Serine is the sole residue subjected to O-linked glycosylation in Acinetobacter baumannii. J Proteome Res. (2024). https://pubs.acs.org/doi/10.1021/acs.jproteome.4c00148 *Corresponding authorship
2 J. M. Lewis, L. Jebeli, P.M.L. Coulon, C.E. Lay, N.E. Scott. Glycoproteomic and Proteomic analysis of Burkholderia cenocepacia reveals glycosylation events within FliF and MotB are dispensable for motility. Microbiology Spectrum (2024) 12(6):e0034624. doi: 10.1128/spectrum.00346-24. *Corresponding authorship
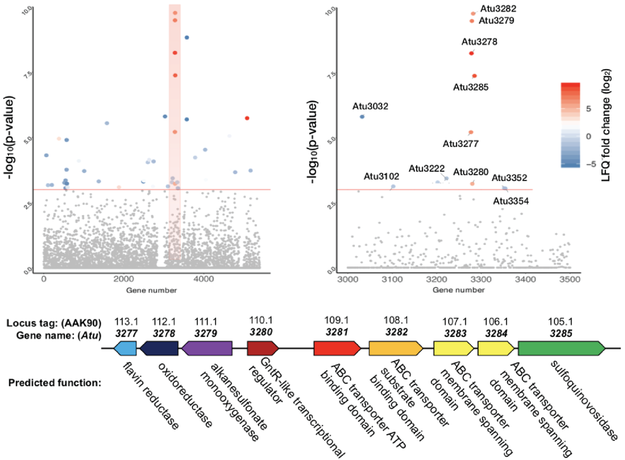
3. I. García-Romero, M. Srivastava, J. Monjarás-Feria, S.O. Korankye, L. MacDonald, N.E. Scott, M.A. Valvano. Drug efflux and lipid A modification by 4-L-aminoarabinose are key mechanisms of polymyxin B resistance in the sepsis pathogen Enterobacter bugandensis. J Glob Antimicrob Resist. (2024) S2213-7165(24)00062-6. DOI: 10.1016/j.jgar.2024.03.012..
4. C. Hunter, T. Derksen, S. Makhsous, M. Doll, S.R. Perez, N.E. Scott, L.M. Willis. Site-specific immobilization of the endosialidase reveals QSOX2 is a novel polysialylated protein. Glycobiology. (2024) cwae026. doi: 10.1093/glycob/cwae026.
5. P.L. Wantuch, C.J. Knoot, L.S. Robinson, E. Vinogradov, N.E. Scott, C.M. Harding, D.A. Rosen. A heptavalent O-antigen bioconjugate vaccine exhibits differential functional antibody responses against diverse Klebsiella pneumoniae isolates. J Infect Dis. (2024) jiae097 doi: 10.1093/infdis/jiae097.
6. K. Sarson-Lawrence, J.M. Hardy, J. Iaria, D. Stockwell, K. Behrens, T. Saiyed, C. Tan, L. Jebeli, N.E. Scott, T. Dite, N.A. Nicola, A. Leis, J.J. Babon, N.J. Kershaw. Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin. Nature Communication (2024) 15(1):1135. doi: 10.1038/s41467-024-45356-2.
7.. E.J. Pardue, M.G. Sartorio; B. Jana, N.E. Scott, W.L Beatty, J.C Ortiz-Marquez, T. van Opijnen, F.F. Hsu, R.F. Potter, M.F Feldman. Dual Membrane-spanning Anti-Sigma Factors Regulate Vesiculation in Bacteroides thetaiotaomicron. Proc Natl Acad Sci U S A. (2024) 121(10) e2321910121. doi: 10.1073/pnas.2321910121.
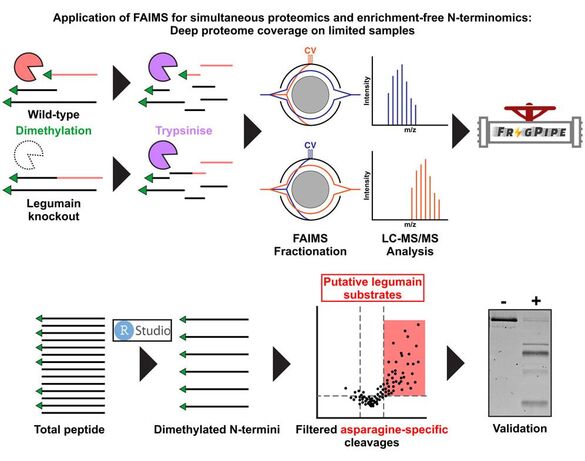
8. A.R. Ziegler, A. Dufour, N.E. Scott*, L.E. Edgington-Mitchell. Ion mobility-based enrichment-free N-terminomics analysis reveals novel legumain substrates in murine spleen. Mol Cell Proteomics. (2024) 23(2):100714. doi: 10.1016/j.mcpro.2024.100714. *Co-Corresponding authorship
3. I. García-Romero, M. Srivastava, J. Monjarás-Feria, S.O. Korankye, L. MacDonald, N.E. Scott, M.A. Valvano. Drug efflux and lipid A modification by 4-L-aminoarabinose are key mechanisms of polymyxin B resistance in the sepsis pathogen Enterobacter bugandensis. J Glob Antimicrob Resist. (2024) S2213-7165(24)00062-6. DOI: 10.1016/j.jgar.2024.03.012..
4. C. Hunter, T. Derksen, S. Makhsous, M. Doll, S.R. Perez, N.E. Scott, L.M. Willis. Site-specific immobilization of the endosialidase reveals QSOX2 is a novel polysialylated protein. Glycobiology. (2024) cwae026. doi: 10.1093/glycob/cwae026.
5. P.L. Wantuch, C.J. Knoot, L.S. Robinson, E. Vinogradov, N.E. Scott, C.M. Harding, D.A. Rosen. A heptavalent O-antigen bioconjugate vaccine exhibits differential functional antibody responses against diverse Klebsiella pneumoniae isolates. J Infect Dis. (2024) jiae097 doi: 10.1093/infdis/jiae097.
6. K. Sarson-Lawrence, J.M. Hardy, J. Iaria, D. Stockwell, K. Behrens, T. Saiyed, C. Tan, L. Jebeli, N.E. Scott, T. Dite, N.A. Nicola, A. Leis, J.J. Babon, N.J. Kershaw. Cryo-EM structure of the extracellular domain of murine Thrombopoietin Receptor in complex with Thrombopoietin. Nature Communication (2024) 15(1):1135. doi: 10.1038/s41467-024-45356-2.
7.. E.J. Pardue, M.G. Sartorio; B. Jana, N.E. Scott, W.L Beatty, J.C Ortiz-Marquez, T. van Opijnen, F.F. Hsu, R.F. Potter, M.F Feldman. Dual Membrane-spanning Anti-Sigma Factors Regulate Vesiculation in Bacteroides thetaiotaomicron. Proc Natl Acad Sci U S A. (2024) 121(10) e2321910121. doi: 10.1073/pnas.2321910121.
8. A.R. Ziegler, A. Dufour, N.E. Scott*, L.E. Edgington-Mitchell. Ion mobility-based enrichment-free N-terminomics analysis reveals novel legumain substrates in murine spleen. Mol Cell Proteomics. (2024) 23(2):100714. doi: 10.1016/j.mcpro.2024.100714. *Co-Corresponding authorship
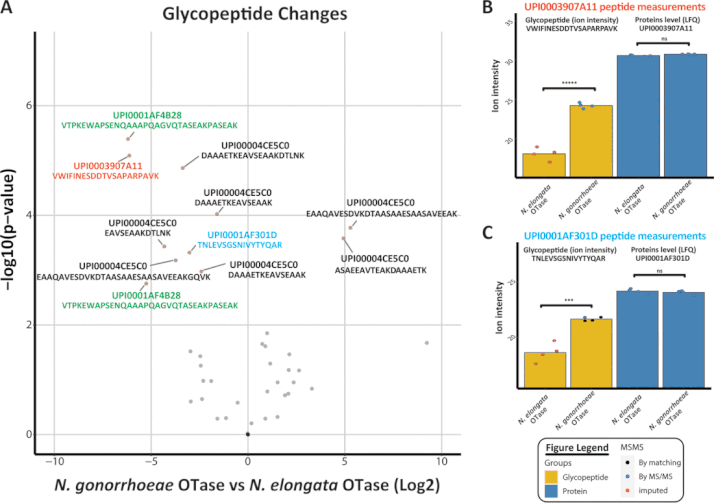
9. C. Hadjineophytou, E. Loh, M. Koomey N.E. Scott*. Combining FAIMS based Glycoproteomics and DIA Proteomics reveals widespread proteome alterations in response to glycosylation occupancy changes in Neisseria gonorrhoeae. Proteomics. (2024) e2300496. doi: 10.1002/pmic.202300496. PMID: 38361220 *Corresponding authorship
2023
10. A. Kaur, I. Pickles, M. Sharma, N.S. Madiedo, N.E. Scott, S. J. Pidot, E. D. Goddard-Borger, G. Davies, S.J. Williams. A widespread family of NAD-dependent sulfoquinovosidases at the gateway to sulfoquinovose catabolism. J. Am. Chem. Soc. (2023) 145(51):28216-28223 doi.org/10.1021/jacs.3c11126..
11. R. K. Loterio, D. R. Thomas, W. Andrade, Y. W. Lee, L. Lima dos Santos, D. P. A. Mascarenhas, T. Maass Steiner, J. Chiaratto, L. F. Fielden, L. Lopes, L. Bird, G.H. Goldman, D. Stojanovski, N. E. Scott, D. S. Zamboni & H. J. Newton. Coxiella co-opts the Glutathione Peroxidase 4 (GPX4) to protect the host cell from oxidative stress-induced cell death. Proc Natl Acad Sci U S A. (2023). 120(36):e2308752120.
12. A. W. Debowski, N.M. Bzdyl, D. R. Thomas, N. E. Scott, C. H. Jenkins, J. Iwasaki, E. A. Kibble, C. A. Khoo, N. J. Scheuplein, P. M. Seibel, T. Lohr, G. Metters, C. S. Bond, I. H. Norville, K. A. Stubbs, N. J. Harmer, Ulrike Holzgrabe, H. J. Newton, M. Sarkar-Tyson. Macrophage infectivity potentiator protein, a peptidyl prolyl cis-trans isomerase, essential for Coxiella burnetii growth and pathogenesis. PLoS pathogens (2023) 19(7):e1011491.
13. J. C. McGuffey, C. D. Jackson-Litteken , G. Di Venanzio , A. A. Zimmer , J. M. Lewis , J. Distel, K. Q Kim , H. Zaher, J. Alfonzo , N. E. Scott, M. F. Feldman. The tRNA methyltransferase TrmB is critical for Acinetobacter baumannii stress responses and pulmonary infection. mBio (2023) e0141623. doi: 10.1128/mbio.01416-23.
14. L. R. Nigos, N. E. Scott, A. G. Brooks, M. Ait-Goughoulte, S. L. Londrigan, P. C. Reading, R. Farrukee. TRIM16 Overexpression in HEK293T Cells Results in Cell Line-Specific Antiviral Activity. Pathogens. (2023) 12(6):852. doi: 10.3390/pathogens12060852.
15. M. G. Sartorio, E. J. Pardue, N. E. Scott, M. F. Feldman. Human gut bacteria tailor extracellular vesicle cargo for the breakdown of diet- and host-derived glycans. Proc Natl Acad Sci U S A. (2023) 120(27):e2306314120. doi: 10.1073/pnas.2306314120.
16. A. Kaur, N. E. Scott, M. Herisse, E. D. Goddard-Borger, S. Pidot, S. J. Williams. Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis. Arch Microbiol. (2023) 205(4):155. doi: 10.1007/s00203-023-03506-y.
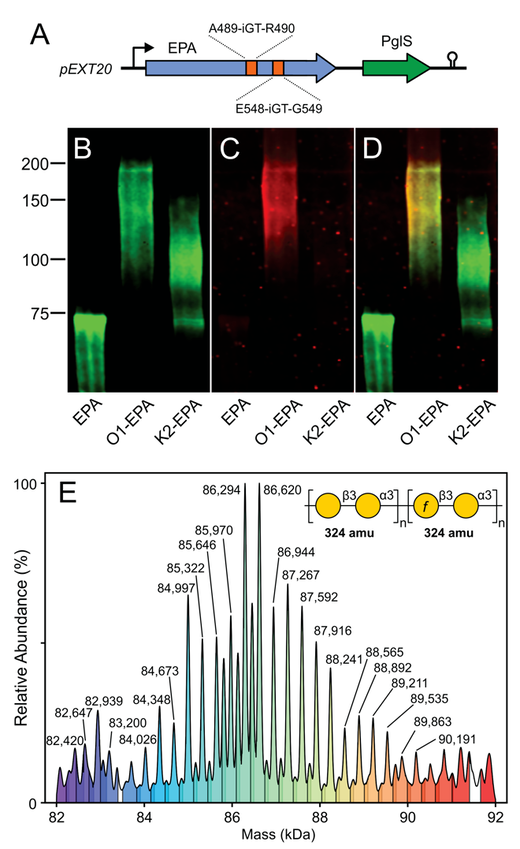
17. P. L. Wantuch, C. J. Knoot, L. S. Robinson, E. Vinogradov, N. E. Scott, C. M. Harding, D. A. Rosen. Capsular polysaccharide inhibits vaccine-induced O-antigen antibody binding and function across both classical and hypervirulent K2:O1 strains of Klebsiella pneumoniae. PLoS pathogens (2023) 19(5):e1011367. doi: 10.1371/journal.ppat.1011367.
11. R. K. Loterio, D. R. Thomas, W. Andrade, Y. W. Lee, L. Lima dos Santos, D. P. A. Mascarenhas, T. Maass Steiner, J. Chiaratto, L. F. Fielden, L. Lopes, L. Bird, G.H. Goldman, D. Stojanovski, N. E. Scott, D. S. Zamboni & H. J. Newton. Coxiella co-opts the Glutathione Peroxidase 4 (GPX4) to protect the host cell from oxidative stress-induced cell death. Proc Natl Acad Sci U S A. (2023). 120(36):e2308752120.
12. A. W. Debowski, N.M. Bzdyl, D. R. Thomas, N. E. Scott, C. H. Jenkins, J. Iwasaki, E. A. Kibble, C. A. Khoo, N. J. Scheuplein, P. M. Seibel, T. Lohr, G. Metters, C. S. Bond, I. H. Norville, K. A. Stubbs, N. J. Harmer, Ulrike Holzgrabe, H. J. Newton, M. Sarkar-Tyson. Macrophage infectivity potentiator protein, a peptidyl prolyl cis-trans isomerase, essential for Coxiella burnetii growth and pathogenesis. PLoS pathogens (2023) 19(7):e1011491.
13. J. C. McGuffey, C. D. Jackson-Litteken , G. Di Venanzio , A. A. Zimmer , J. M. Lewis , J. Distel, K. Q Kim , H. Zaher, J. Alfonzo , N. E. Scott, M. F. Feldman. The tRNA methyltransferase TrmB is critical for Acinetobacter baumannii stress responses and pulmonary infection. mBio (2023) e0141623. doi: 10.1128/mbio.01416-23.
14. L. R. Nigos, N. E. Scott, A. G. Brooks, M. Ait-Goughoulte, S. L. Londrigan, P. C. Reading, R. Farrukee. TRIM16 Overexpression in HEK293T Cells Results in Cell Line-Specific Antiviral Activity. Pathogens. (2023) 12(6):852. doi: 10.3390/pathogens12060852.
15. M. G. Sartorio, E. J. Pardue, N. E. Scott, M. F. Feldman. Human gut bacteria tailor extracellular vesicle cargo for the breakdown of diet- and host-derived glycans. Proc Natl Acad Sci U S A. (2023) 120(27):e2306314120. doi: 10.1073/pnas.2306314120.
16. A. Kaur, N. E. Scott, M. Herisse, E. D. Goddard-Borger, S. Pidot, S. J. Williams. Identification of levoglucosan degradation pathways in bacteria and sequence similarity network analysis. Arch Microbiol. (2023) 205(4):155. doi: 10.1007/s00203-023-03506-y.
17. P. L. Wantuch, C. J. Knoot, L. S. Robinson, E. Vinogradov, N. E. Scott, C. M. Harding, D. A. Rosen. Capsular polysaccharide inhibits vaccine-induced O-antigen antibody binding and function across both classical and hypervirulent K2:O1 strains of Klebsiella pneumoniae. PLoS pathogens (2023) 19(5):e1011367. doi: 10.1371/journal.ppat.1011367.
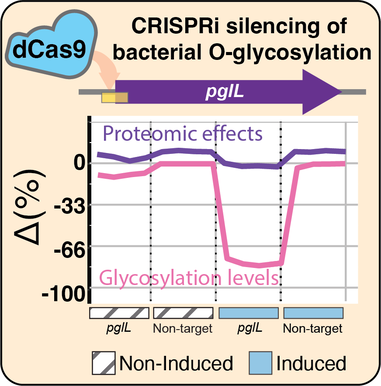
18. J. M. Lewis, N. E. Scott*. CRISPRi mediated silencing of Burkholderia O-linked glycosylation systems enables the depletion of glycosylation yet results in modest proteome impacts. J Proteome Res. (2023) 22(6):1762-1778. doi: 10.1021/acs.jproteome.2c00790. *Corresponding authorship
19. A. John, S.M. Bader, N.M Soler, K. Wiradiputri, S. Tichkule, S.T. Smyth, S.A. Ralph, A.R. Jex, N.E. Scott*, C.J. Tonkin*, E.D. Goddard-Borger*. Conservation, abundance, glycosylation profile, and localization of the TSP protein family in Cryptosporidium parvum. J Biol Chem. 2023 Feb 10:103006. doi: 10.1016/j.jbc.2023.103006. * Co-Corresponding authorship
20. J.S. Bloch, A. John, R. Mao, S. Mukherjee, J. Boilevin, R.N. Irobalieva, T. Darbre, N.E. Scott, J.L Reymond5, A.A. Kossiakoff, E.D.Goddard-Borger, K.P. Locher. Structure, sequon binding, donor recruitment, and mechanism of tryptophan C-mannosyltransferase Nature Chemical Biology (2023). doi: 10.1038/s41589-022-01219-9
21. C.J. Knoot, P.L. Wantuch, L.S. Robinson, D.A. Rosen, N.E. Scott, C.M. Harding. Discovery and characterization of a new class of O-linking oligosaccharyltransferases from the Moraxellaceae family. Glycobiology. (2023) Oct 14:cwac070. doi: 10.1093/glycob/cwac070.
22. J.B. McDonald, N.E. Scott, G.J. Underwood, D.M. Andrews, T.T.H. Van, R.J. Moore. Characterisation of N-linked protein glycosylation in the bacterial pathogen Campylobacter hepaticus. Sci Rep. (2023) 13(1):227. doi: 10.1038/s41598-022-26532-0.
20. J.S. Bloch, A. John, R. Mao, S. Mukherjee, J. Boilevin, R.N. Irobalieva, T. Darbre, N.E. Scott, J.L Reymond5, A.A. Kossiakoff, E.D.Goddard-Borger, K.P. Locher. Structure, sequon binding, donor recruitment, and mechanism of tryptophan C-mannosyltransferase Nature Chemical Biology (2023). doi: 10.1038/s41589-022-01219-9
21. C.J. Knoot, P.L. Wantuch, L.S. Robinson, D.A. Rosen, N.E. Scott, C.M. Harding. Discovery and characterization of a new class of O-linking oligosaccharyltransferases from the Moraxellaceae family. Glycobiology. (2023) Oct 14:cwac070. doi: 10.1093/glycob/cwac070.
22. J.B. McDonald, N.E. Scott, G.J. Underwood, D.M. Andrews, T.T.H. Van, R.J. Moore. Characterisation of N-linked protein glycosylation in the bacterial pathogen Campylobacter hepaticus. Sci Rep. (2023) 13(1):227. doi: 10.1038/s41598-022-26532-0.
2022
23. G. Polanco, N.E. Scott, L.F. Lye, S.M. Beverley. Expanded Proteomic Survey of the Human Parasite Leishmania major Focusing on Changes in Null Mutants of the Golgi GDP-Mannose/Fucose/Arabinopyranose Transporter LPG2 and of the Mitochondrial Fucosyltransferase FUT1. Microbiol Spectr. (2022); 10(6):e0305222. doi: 10.1128/spectrum.03052-22.
24. J. Aceil, A.V. Paschall, C.J. Knoot, L.S. Robinson, N.E. Scott, M.F. Feldman, C.M. Harding, F.Y. Avci. Immunogenicity and Protective Efficacy of a Prototype Pneumococcal Bioconjugate Vaccine. Vaccine (2022) 40(42):6107-6113. doi: 10.1016/j.vaccine.2022.09.018
25. S. Lopaticki, R. McConville, A. John, N. Geoghegan, S. Deen Mohamed, L. Verzier, R. W. J. Steel, C. Evelyn, M. T. O’Neill, N. M. Soler, N. E. Scott, K. L. Rogers, E. D. Goddard-Borger, J. A. Boddey. Tryptophan C-mannosylation is critical for Plasmodium falciparum transmission. Nature Communication (2022) 13, 4400. doi: 10.1038/s41467-022-32076-8.
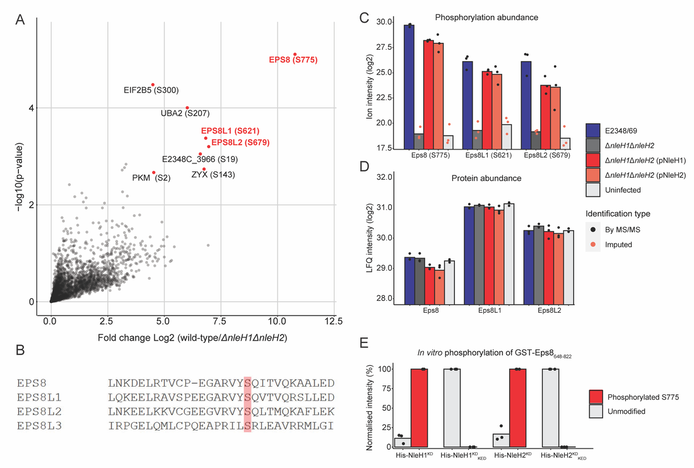
26. G. L. Pollock, A. M. Grishin, C. Giogha, J. Gan, C. Oates, P. J. McMillan, I. Gaeta, M.J. Tyska, J.S. Pearson, N. E. Scott, M. Cygler, E. L. Hartland. Targeting of microvillus protein Eps8 by the NleH effector kinases from enteropathogenic E. coli. PNAS (2022) 119, e2204332119. doi: 10.1073/pnas.2204332119
24. J. Aceil, A.V. Paschall, C.J. Knoot, L.S. Robinson, N.E. Scott, M.F. Feldman, C.M. Harding, F.Y. Avci. Immunogenicity and Protective Efficacy of a Prototype Pneumococcal Bioconjugate Vaccine. Vaccine (2022) 40(42):6107-6113. doi: 10.1016/j.vaccine.2022.09.018
25. S. Lopaticki, R. McConville, A. John, N. Geoghegan, S. Deen Mohamed, L. Verzier, R. W. J. Steel, C. Evelyn, M. T. O’Neill, N. M. Soler, N. E. Scott, K. L. Rogers, E. D. Goddard-Borger, J. A. Boddey. Tryptophan C-mannosylation is critical for Plasmodium falciparum transmission. Nature Communication (2022) 13, 4400. doi: 10.1038/s41467-022-32076-8.
26. G. L. Pollock, A. M. Grishin, C. Giogha, J. Gan, C. Oates, P. J. McMillan, I. Gaeta, M.J. Tyska, J.S. Pearson, N. E. Scott, M. Cygler, E. L. Hartland. Targeting of microvillus protein Eps8 by the NleH effector kinases from enteropathogenic E. coli. PNAS (2022) 119, e2204332119. doi: 10.1073/pnas.2204332119
27. I. Bagdonaite, S.A. Malaker, D.A. Polasky, N.M. Riley, K. Schjoldager, S.Y. Vakhrushev A. Halim K.F. Aoki-Kinoshita, A.I. Nesvizhskii, C.R. Bertozzi, H.H. Wandall, B.L. Parker, M. Thaysen-Andersen, N.E. Scott. Glycoproteomics. Nature Reviews Methods Primers (2022) DOI: 10.1038/s43586-022-00128-4
28. C. D. Jackson-Litteken, G. Di Venanzio , N.H. Le , N.E. Scott , B. Djahanschiri , J.S. Distel , E.J. Pardue , I. Ebersberger , M.F. Feldman. InvL, an Invasin-Like Adhesin, Is a Type II Secretion System Substrate Required for Acinetobacter baumannii Uropathogenesis. mBio (2022) 13(3):e0025822. doi: 10.1128/mbio.00258-22
29. C. Hadjineophytou, J.H. Anonsen, T. Svingerud, T.D. Mortimer, Y.H. Grad, N.E. Scott, M. Koomey. Sculpting the Bacterial O-Glycoproteome: Functional Analyses of Orthologous Oligosaccharyltransferases with Diverse Targeting Specificities. mBio (2022) https://doi.org/10.1128/mbio.03797-21
29. C. Hadjineophytou, J.H. Anonsen, T. Svingerud, T.D. Mortimer, Y.H. Grad, N.E. Scott, M. Koomey. Sculpting the Bacterial O-Glycoproteome: Functional Analyses of Orthologous Oligosaccharyltransferases with Diverse Targeting Specificities. mBio (2022) https://doi.org/10.1128/mbio.03797-21
30. S. El Qaidi, N.E. Scott, P.R. Hardwidge. Arginine glycosylation regulates UDP-GlcNAc biosynthesis in Salmonella enterica. Scientific Reports (2022) 12(1):5293. doi: 10.1038/s41598-022-09276-9
31. A.J. Hooppaw , J.C. McGuffey , G.Di Venanzio , J.C. Ortiz-Marquez , B.S. Weber , T.J. Lightly , T. van Opijnen , N.E. Scott , S.T. Cardona , M.F. Feldman The Phenylacetic Acid Catabolic Pathway Regulates Antibiotic and Oxidative Stress Responses in Acinetobacter. mBio (2022) https://doi.org/10.1128/mbio.01863-21
32. M. Sharma, J.P. Lingford, M. Petricevic, A. Snow, Y. Zhang, M. Jarva, J. W.Y. Mui, N.E. Scott, E.C. Saunders, R. Mao, R. Epa, B. Moiera, D. Ascher, M.J. McConville, G.J. Davies, S.J. Williams, and E.D. Goddard-Borger. Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria. Proc Natl Acad Sci U S A. (2022) 119(4):e2116022119. doi: 10.1073/pnas.2116022119. PMID: 35074914
31. A.J. Hooppaw , J.C. McGuffey , G.Di Venanzio , J.C. Ortiz-Marquez , B.S. Weber , T.J. Lightly , T. van Opijnen , N.E. Scott , S.T. Cardona , M.F. Feldman The Phenylacetic Acid Catabolic Pathway Regulates Antibiotic and Oxidative Stress Responses in Acinetobacter. mBio (2022) https://doi.org/10.1128/mbio.01863-21
32. M. Sharma, J.P. Lingford, M. Petricevic, A. Snow, Y. Zhang, M. Jarva, J. W.Y. Mui, N.E. Scott, E.C. Saunders, R. Mao, R. Epa, B. Moiera, D. Ascher, M.J. McConville, G.J. Davies, S.J. Williams, and E.D. Goddard-Borger. Oxidative desulfurization pathway for complete catabolism of sulfoquinovose by bacteria. Proc Natl Acad Sci U S A. (2022) 119(4):e2116022119. doi: 10.1073/pnas.2116022119. PMID: 35074914
33. C.S. Ang, J. Sacharz, M.G Leeming, S. Nie, S. Varshney, N.E Scott, N.A Williamson. Getting more out of FLAG-Tag co-immunoprecipitation mass spectrometry experiments using FAIMS. J Proteomics. (2022) 254:104473. doi: 10.1016/j.jprot.2021.104473 PMID: 34990820
34. P.D. Veith, M. Shoji, N.E Scott, E.C. Reynolds. Characterisation of the O-glycoproteome of Porphyromonas gingivalis. Microbiol. Spectr. (2022) 10(1):e0150221. PMID: 34985300
34. P.D. Veith, M. Shoji, N.E Scott, E.C. Reynolds. Characterisation of the O-glycoproteome of Porphyromonas gingivalis. Microbiol. Spectr. (2022) 10(1):e0150221. PMID: 34985300
2021
35. J.M. Lewis, P.M.L. Coulon, T.A. McDaniels, N.E. Scott. The Application of Open Searching-based Approaches for the Identification of Acinetobacter baumannii O-linked Glycopeptides. J Vis Exp. 2021; (177). PMID: 34806714
36. R. Kawahara, A.C, K. Alagesan, M. Bern, W. Cao, R.J. Chalkley, K. Cheng, M.S. Choo, N. Edwards, R. Goldman, M. Hoffmann, Y. Hu, Y. Huang, J.Y Kim, D. Kletter, B. Liquet-Weiland, M. Liu, Y. Mechref, B Meng, S. Neelamegham, T. Nguyen-Khuong, J. Nilsson, A. Pap, G.W Park, B.L. Parker, C.L. Pegg, J.M. Penninger, T.K. Phung, M. Pioch, E. Rapp, E. Sakalli, M. Sanda, B.L. Schulz, N.E. Scott, G. Sofronov, J. Stadlmann, S.Y. Vakhrushev, C.M. Woo, H.Y Wu, P. Yang, W. Ying, H. Zhang, Y. Zhang, J. Zhao, J. Zaia, S.M. Haslam, G. Palmisano, J.S Yoo, G. Larson, K.H Khoo, K.F. Medzihradszky, D. Kolarich, N.H. Packer and M. Thaysen-Andersen. Community evaluation of glycoproteomics informatics solutions reveals high-performance search strategies of serum N- and O-glycopeptide data. Nat Methods. 18(11):1304-1316. PMID: 34725484
37. J.A. Duke, A.V. Paschall, L.S. Robinson, C.J. Knoot, E. Vinogradov, N.E. Scott, M.F. Feldman, F. Y. Avci, C.M. Harding. Development and Immunogenicity of a Prototype Multivalent Group B Streptococcus Bioconjugate Vaccine. ACS Infect Dis. (2021) doi: 10.1021/acsinfecdis.1c00415. PMID: 34633812
38. P.D. Veith, N.E Scott, E.C. Reynolds. Characterization of the O-Glycoproteome of Tannerella forsythia. mSphere (2021) e0064921. PMID: 34523981
39. A.J. Hayes, J.M. Lewis, M.R. Davies, N.E. Scott*. Burkholderia PglL enzymes are Serine preferring oligosaccharyltransferases which target conserved proteins across the Burkholderia genus. Commun Biol 4, 1045 (2021). PMID: 34493791 *Corresponding authorship
37. J.A. Duke, A.V. Paschall, L.S. Robinson, C.J. Knoot, E. Vinogradov, N.E. Scott, M.F. Feldman, F. Y. Avci, C.M. Harding. Development and Immunogenicity of a Prototype Multivalent Group B Streptococcus Bioconjugate Vaccine. ACS Infect Dis. (2021) doi: 10.1021/acsinfecdis.1c00415. PMID: 34633812
38. P.D. Veith, N.E Scott, E.C. Reynolds. Characterization of the O-Glycoproteome of Tannerella forsythia. mSphere (2021) e0064921. PMID: 34523981
39. A.J. Hayes, J.M. Lewis, M.R. Davies, N.E. Scott*. Burkholderia PglL enzymes are Serine preferring oligosaccharyltransferases which target conserved proteins across the Burkholderia genus. Commun Biol 4, 1045 (2021). PMID: 34493791 *Corresponding authorship
40. G. Sycz, G. Di Venanzio, M.G. Sartorio, L. Nguyen-Hung, N.E. Scott, W.L. Beatty, M.F. Feldman. Modern Acinetobacter baumannii clinical isolates replicate inside spacious vacuoles and egress from macrophages. PLOS pathogens (2021) 17(8):e1009802. PMID: 34370792
41 R. Mao, S. Xi, S. Shah, M.J. Roy, A. John, J.P. Lingford, G. Gäde, N.E. Scott, E.D. Goddard-Borger. Synthesis of C-Mannosylated Glycopeptides Enabled by Ni-Catalyzed Photoreductive Cross-Coupling Reactions. J Am Chem Soc. (2021)
42. R. Grinter, F.C. Morris, R.A. Dunstan, P.M. Leung, A. Kropp, M. Belousoff, S.D. Gunasinghe, N.E. Scott, S. Beckham, A.Y. Peleg, C. Greening, J. Li, E. Heinz, T. Lithgow. BonA from Acinetobacter baumannii forms a divisomelocalized decamer that supports outer envelope function. mBio (2021) 12:e01480-21.
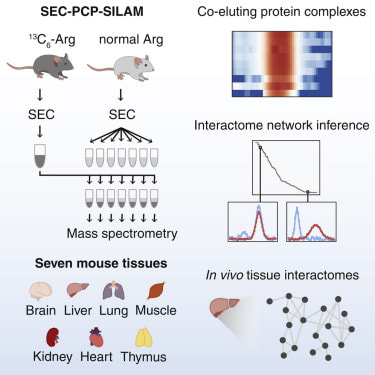
43. M.A. Skinnider, N.E. Scott*, A. Prudova, Craig H. Kerr, N. Stoynov, R.G. Stacey, Q.W.T. Chan, D. Rattray, J. Gsponer and L.J. Foster. An atlas of protein-protein interactions across mouse tissues. Cell (2021) DOI:https://doi.org/10.1016/j.cell.2021.06.003 *Joint lead author
44. C. Giogha*, N.E Scott,* T. Wong Fok Lung, G.L Pollock, M. Harper, E.D Goddard-Borger, J.S. Pearson and E.L. Hartland. NleB2 from enteropathogenic Escherichia coli is a novel arginine-glucose transferase effector. PLoS Pathogen (2021) 17(6):e1009658. *Joint lead author
45. P. Pymm, A. Adair, L.J. Chan, J.P. Cooney, F.L. Mordant, C.C. Allison, E. Lopez, E.R. Haycroft, M.T. O'Neill, L.L. Tan, M.H. Dietrich, D. Drew, M. Doerflinger, M.A. Dengler, N.E. Scott, A.K. Wheatley, N.A. Gherardin, H. Venugopal, D. Cromer, M.P. Davenport, R. Pickering, D.I. Godfrey, D.F.J. Purcell, S.J. Kent, A.W. Chung, K. Subbarao, M. Pellegrini, A. Glukhova, W.H. Tham. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc Natl Acad Sci U S A. (2021), 118(19):e2101918118.
46. D.R. Thomas and N.E. Scott. Glycoproteomics: growing up fast. Current Opinion in Structural Biology (2021) 68, pp. 18–25
47. N.E. Scott and L.E. Edgington-Mitchell. Editorial overview: Systems biology and the rise and rise of omics approaches. (2021) Current Opinion in Chemical Biology. PMID: 33419639
48. S. El Qaidi, N.E. Scott and P.R. Hardwidge. Arginine glycosylation enhances methylglyoxal detoxification. Scientific Reports. (2021) 11(1):3834. PMID: 33589708
49. A. John, M.A. Järvå, S. Shah, R. Mao, S Chappaz, R.W. Birkinshaw, P.E. Czabotar, A.W. Lo, N.E. Scott and E.D. Goddard-Borger. Yeast- and antibody-based tools for studying tryptophan C-mannosylation. Nature Chemical Biology. (2021) doi: 10.1038/s41589-020-00727-w. PMID: 33542533
46. D.R. Thomas and N.E. Scott. Glycoproteomics: growing up fast. Current Opinion in Structural Biology (2021) 68, pp. 18–25
47. N.E. Scott and L.E. Edgington-Mitchell. Editorial overview: Systems biology and the rise and rise of omics approaches. (2021) Current Opinion in Chemical Biology. PMID: 33419639
48. S. El Qaidi, N.E. Scott and P.R. Hardwidge. Arginine glycosylation enhances methylglyoxal detoxification. Scientific Reports. (2021) 11(1):3834. PMID: 33589708
49. A. John, M.A. Järvå, S. Shah, R. Mao, S Chappaz, R.W. Birkinshaw, P.E. Czabotar, A.W. Lo, N.E. Scott and E.D. Goddard-Borger. Yeast- and antibody-based tools for studying tryptophan C-mannosylation. Nature Chemical Biology. (2021) doi: 10.1038/s41589-020-00727-w. PMID: 33542533
50. G. Wang, L. Glaser, N.E. Scott, Y.F. Mohamed, R. Ingram, K. Laroucau and M.A. Valvano. A glycoengineered antigen exploiting a conserved protein O-glycosylation pathway in the Burkholderia genus for detection of glanders infections. Virulence. (2021) 12(1):493-506. PMID: 33509023
51. C.J. Kearney, S.J. Vervoort, K.M. Ramsbottom, I Todorovski, E.J. Lelliott, M. Zethoven, L. Pijpers, B.P. Martin, T. Semple, L. Martelotto, J.A. Trapani, I.A. Parish, N.E. Scott, J. Oliaro and R.W. Johnstone. SUGAR-seq enables simultaneous detection of glycans, epitopes, and the transcriptome in single cells. Science Advances (2021) 19;7(8):eabe3610. PMID: 33608275
52. G. Deliyannis, C.Y. Wong, H.A. McQuilten, A. Bachem, M.V. Clarke, X. Jia, K. Horrocks, W. Zeng, J. Girkin, N.E. Scott, S.L. Londrigan, P.C. Reading, N.W. Bartlett, K. Kedzierska, L.E. Brown, F.A. Mercuri, C. Demaison, D.C. Jackson and B.Y. Chua. TLR-2-mediated activation of innate responses in the upper airways confers anti-viral protection of the lungs. JCI Insight. (2021) 9:140267. doi: 10.1172/jci.insight.140267. PMID: 33561017
53. A. R. Ahmad Izaham C.S. Ang, S. Nie, L.E. Bird, N.A. Williamson and N. E. Scott. What are we missing by using hydrophilic enrichment? Improving bacterial glycoproteome coverage using total proteome and FAIMS analysis. J Proteome Res. (2021) 20(1):599-612 PMID: 33125241.
51. C.J. Kearney, S.J. Vervoort, K.M. Ramsbottom, I Todorovski, E.J. Lelliott, M. Zethoven, L. Pijpers, B.P. Martin, T. Semple, L. Martelotto, J.A. Trapani, I.A. Parish, N.E. Scott, J. Oliaro and R.W. Johnstone. SUGAR-seq enables simultaneous detection of glycans, epitopes, and the transcriptome in single cells. Science Advances (2021) 19;7(8):eabe3610. PMID: 33608275
52. G. Deliyannis, C.Y. Wong, H.A. McQuilten, A. Bachem, M.V. Clarke, X. Jia, K. Horrocks, W. Zeng, J. Girkin, N.E. Scott, S.L. Londrigan, P.C. Reading, N.W. Bartlett, K. Kedzierska, L.E. Brown, F.A. Mercuri, C. Demaison, D.C. Jackson and B.Y. Chua. TLR-2-mediated activation of innate responses in the upper airways confers anti-viral protection of the lungs. JCI Insight. (2021) 9:140267. doi: 10.1172/jci.insight.140267. PMID: 33561017
53. A. R. Ahmad Izaham C.S. Ang, S. Nie, L.E. Bird, N.A. Williamson and N. E. Scott. What are we missing by using hydrophilic enrichment? Improving bacterial glycoproteome coverage using total proteome and FAIMS analysis. J Proteome Res. (2021) 20(1):599-612 PMID: 33125241.
2020
54. L.F. Fielden, N.E. Scott, C.S. Palmer, C.A. Khoo, H.J. Newton, D. Stojanovski. Proteomic identification of Coxiella burnetii effector proteins targeted to the host cell mitochondria during infection. Mol Cell Proteomics. (2020) mcp.RA120.002370. PMID: 33177156
55. M.F. Haurat, N.E. Scott, G. Di Venanzio, J. Lopez, B. Pluvinage, A.B. Boraston, M.J. Ferracane, M.F. Feldman. The Glycoprotease CpaA Secreted by Medically Relevant Acinetobacter Species Targets Multiple O-Linked Host Glycoproteins. mBio. (2020) 11(5). pii: e02033-20. PMID: 33024038
56. J. Gan, N.E. Scott, J.P.M. Newson, R.R. Wibawa, T. Wong Fok Lung, G.L. Pollock, G.Z. Ng, I. van Driel, J.S. Pearson, E.L. Hartland, C. Giogha. The Salmonella Effector SseK3 Targets Small Rab GTPases. Front Cell Infect Microbiol. (2020) 10:419 PMID: 32974215
57. J.A Juno, H.- Tan, W.S Lee, A. Reynaldi, H.G. Kelly, K. Wragg, R. Esterbauer, H.E. Kent, C.J. Batten, F.L. Mordant, N.A. Gherardin, P. Pymm, M.H. Dietrich, N.E. Scott, W.H. Tham, D.I. Godfrey, K. Subbarao, M.P. Davenport, S.J. Kent, A.K. Wheatley. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nature Medicine. (2020) 26(9):1428-1434. PMID: 32661393
58. A. R. Ahmad Izaham, N. E. Scott. Open database searching enables the identification and comparison of bacterial glycoproteomes without defining glycan compositions prior to searching. Mol Cell Proteomics. (2020) PMID: 32576591.
55. M.F. Haurat, N.E. Scott, G. Di Venanzio, J. Lopez, B. Pluvinage, A.B. Boraston, M.J. Ferracane, M.F. Feldman. The Glycoprotease CpaA Secreted by Medically Relevant Acinetobacter Species Targets Multiple O-Linked Host Glycoproteins. mBio. (2020) 11(5). pii: e02033-20. PMID: 33024038
56. J. Gan, N.E. Scott, J.P.M. Newson, R.R. Wibawa, T. Wong Fok Lung, G.L. Pollock, G.Z. Ng, I. van Driel, J.S. Pearson, E.L. Hartland, C. Giogha. The Salmonella Effector SseK3 Targets Small Rab GTPases. Front Cell Infect Microbiol. (2020) 10:419 PMID: 32974215
57. J.A Juno, H.- Tan, W.S Lee, A. Reynaldi, H.G. Kelly, K. Wragg, R. Esterbauer, H.E. Kent, C.J. Batten, F.L. Mordant, N.A. Gherardin, P. Pymm, M.H. Dietrich, N.E. Scott, W.H. Tham, D.I. Godfrey, K. Subbarao, M.P. Davenport, S.J. Kent, A.K. Wheatley. Humoral and circulating follicular helper T cell responses in recovered patients with COVID-19. Nature Medicine. (2020) 26(9):1428-1434. PMID: 32661393
58. A. R. Ahmad Izaham, N. E. Scott. Open database searching enables the identification and comparison of bacterial glycoproteomes without defining glycan compositions prior to searching. Mol Cell Proteomics. (2020) PMID: 32576591.
59. J. Li, R. Epa, N.E. Scott, D. Skoneczny, M. Sharma, A.J.D. Snow, J.P. Lingford, E.D. Goddard-Borger, G.J. Davies, M.J. McConville, S.J. Williams. A sulfoglycolytic Entner-Doudoroff pathway in Rhizobium leguminosarum bv. trifolii SRDI565. Applied Environmental Microbiology. (2020) 86(15). pii: e00750-20 PMID: 32444469
60. J. Duma, H. Nothaft, D. Weaver, C. Fodor, B. Beadle, D. Linton, S. Benoit, N.E. Scott, R. Maier, C.M. Szymanski. Influence of protein glycosylation on Campylobacter fetus physiology. Frontiers in Microbiology. (2020) 11:1191. PMID: 32625174
61. M. Järvå, J.P. Lingford, A. John, N. Madiedo Soler N.E. Scott, E.D. Goddard-Borger. Trefoil factors share a lectin activity that defines their role in mucus. Nat Commun. (2020) 11, 2265
62. Z. Fan*, J.R. Devlin*, S. Hogg, M.A. Doyle, P.F. Harrison, I. Todorovski, L.A. Cluse, D. Knight, J. Sandow, G. Gregory, A. Fox, T.H. Beilharz, N. Kwiatkowski, N.E. Scott, A. Tufegdzic-Vidacovic, G. Kelly, J. Svejstrup, M. Geyer, N. Gray, S.J. Vervoort, R.W. Johnstone. CDK13 cooperates with CDK12 to control global RNA polymerase II processivity. Science Advances (2020) 6(18), eaaz5041
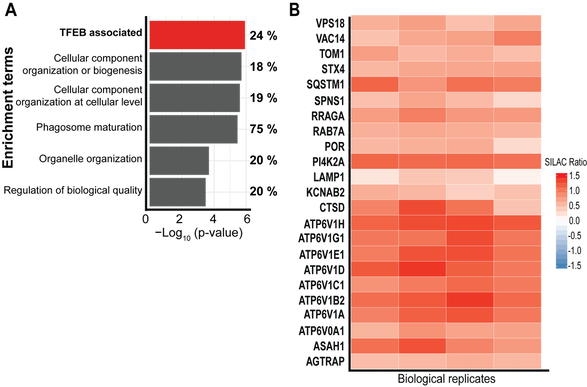
63. B. Padmanabhan, L.F. Fielden, A. Hachani, P. Newton, D.R. Thomas, H.J. Cho, C.A. Khoo, D. Stojanovski, C.R. Roy, N.E. Scott, H.J. Newton. Biogenesis of the spacious Coxiella-containing vacuole depends on host transcription factors TFEB and TFE3. Infect Immun. (2020) 20;88(3):e00534-19
64. S. El Qaidi, N.E. Scott, M.P. Hays, B.V. Geisbrecht, S. Watkins, P.R. Hardwidge. An intra-bacterial activity for a T3SS effector. Scientific Reports (2020), 10 (1), 1073
2019
65. C.C. Oppy, L. Jebeli, M. Kuba, C.V. Oates, R Strugnell, L.E. Edgington-Mitchell, M.A. Valvano, E.L. Hartland, H.J. Newton, N.E. Scott. Loss of O-Linked Protein Glycosylation in Burkholderia cenocepacia Impairs Biofilm Formation and Siderophore Activity and Alters Transcriptional Regulators. mSphere. (2019) Nov 13;4(6). pii: e00660-19
66. E. Lopez, N.E. Scott, B.D. Wines, P.M Hogarth, A. K. Wheatley, S. J. Kent and A.W. Chung. Low pH Exposure During Immunoglobulin G Purification Methods Results in Aggregates That Avidly Bind Fcγ Receptors: Implications for Measuring Fc Dependent Antibody Functions. Front Immunol. (2019). 10:2415
67. M.T Moutaoufik, R. Malty, S. Amin, Q. Zhang, S. Phanse, A. Gagarinova, M. Zilocchi, L. Hoell, Z. Minic, M. Gagarinova, H Aoki, J. Stockwell, M. Jessulat, F. Goebels, K. Broderick, N.E. Scott, J. Vlasblom, G. Musso, B. Prasad, E. Lamantea, B. Garavaglia, A. Rajput, K. Murayama, Y. Okazaki, L.J. Foster, G.D. Bader, F.S. Cayabyab, M. Babu. Rewiring of the Human Mitochondrial Interactome during Neuronal Reprogramming Reveals Regulators of the Respirasome and Neurogenesis. iScience. (2019) 19:1114-1132. PMID: 31536960
68. M.L Carlson, R.G. Stacey, J.W. Young, I.S. Wason, Z. Zhao, D.G. Rattray, N. Scott, C.H. Kerr, M. Babu, L.J. Foster, F. Duong Van Hoa. Profiling the Escherichia coli membrane protein interactome captured in Peptidisc libraries. Elife. (2019) Jul 31;8.
69. M.F Feldman, A.E. Mayer Bridwell, N.E. Scott, E. Vinogradov, S.R. McKee, S.M. Chavez, J. Twentyman, C.L. Stallings, D.A. Rosen, C.M. Harding. A promising bioconjugate vaccine against hypervirulent Klebsiella pneumoniae. Proc Natl Acad Sci U S A. (2019) 116(37):18655-18663
70. Y.F. Mohamed*, N.E. Scott*, A. Molinaro, C. Creuzenet, X Ortega, G. Lertmemongkolchai, M.M. Tunney, H. Green, A.M. Jones, D. DeShazer, B.J. Currie, L.J. Foster, R. Ingram, C. De Castro and M.A. Valvano. A general protein O-glycosylation machinery conserved in Burkholderia species improves bacterial fitness and elicits glycan immunogenicity in humans. J Biol Chem (2019) 294(36):13248-13268 *Joint lead author
71. N.M. Bzdyl, N.E. Scott, I.H. Norville, A.E. Scott, T.Atkins, S.Pang, D.S. Sarovich, G. Coombs, T.J.J. Inglis, C.M. Kahler and M. Sarkar-Tyson.. Peptidyl-prolyl isomerase, ppiB, is essential for proteome homeostasis and virulence in Burkholderia pseudomallei . Infect Immun. (2019) 87(10). pii: e00528-19.
72. N.E. Scott. Expanding our understanding of the role of microbial glycoproteomes through high-throughput mass spectrometry approaches. Glycoconj J. (2019) Jul 4
73. G Di Venanzio, A. L. Flores-Mireles, J.J. Calix, M.F. Haurat, N.E. Scott, L.D. Palmer, R.F. Potter, M.E. Hibbins, L. Friedman, B. Wang, G. Dantas, E.P. Skaar, S.J. Hultgren, M.F. Feldman. Urinary tract colonization is enhanced by an endogenous plasmid that regulates chromosomal genes in uropathogenic Acinetobacter baumannii. Nat Commun. (2019) 24;10(1):2763.
74. J. Newson*, N.E. Scott*, I.Y.W. Chung, T.W. Fok Lung, C. Giogha, N. Wang, R. Strugnell, N. Brown, M. Cygler, J. Pearson, E. Hartland. Salmonella effectors SseK1 and SseK3 target death domain proteins in the TNF and TRAIL signaling pathways. Mol. Cell Proteomics (2019) 18(6):1138-1156. *Joint lead author
75. C.M Harding, M.A. Nasr MA, N.E. Scott, G. Goyette-Desjardins, H. Nothaft, A.E. Mayer, S.M. Chavez, J.P Huynh, R.L Kinsella, C.M Szymanski, C.L Stallings, M Segura, M.F Feldman. A platform for glycoengineering a polyvalent pneumococcal bioconjugate vaccine using E. coli as a host. Nat Commun. (2019) Feb 21;10(1):891.
76. J.A Cain, A.L Dale, P. Niewold, W.P Klare, L Man, M.Y White, N.E Scott, S.J Cordwell. Proteomics reveals multiple phenotypes associated with N-linked glycosylation in Campylobacter jejuni. Mol Cell Proteomics. (2019). 18(4):715-734
77. S.L Vogt, R Scholz, Y Peng, R.L Guest, N.E Scott, S.E Woodward, L.J Foster, T.L Raivio, B.B Finlay. Characterization of the Citrobacter rodentium Cpx regulon and its role in host infection. Mol Microbiol. (2019) 111(3):700-716.
78. S. Khurana, M.J. Coffey, A. John, A.D. Uboldi, M.H. Huynh, R.J. Stewart, V.B Carruthers, C. Tonkin, E.D Goddard-Borger, N.E Scott#. Protein O-fucosyltransferase 2–mediated O-glycosylation of the adhesin MIC2 is dispensable for Toxoplasma gondii tachyzoite infection. J Biol Chem (2019) 1;294(5):1541-1553. # corresponding authorship
72. N.E. Scott. Expanding our understanding of the role of microbial glycoproteomes through high-throughput mass spectrometry approaches. Glycoconj J. (2019) Jul 4
73. G Di Venanzio, A. L. Flores-Mireles, J.J. Calix, M.F. Haurat, N.E. Scott, L.D. Palmer, R.F. Potter, M.E. Hibbins, L. Friedman, B. Wang, G. Dantas, E.P. Skaar, S.J. Hultgren, M.F. Feldman. Urinary tract colonization is enhanced by an endogenous plasmid that regulates chromosomal genes in uropathogenic Acinetobacter baumannii. Nat Commun. (2019) 24;10(1):2763.
74. J. Newson*, N.E. Scott*, I.Y.W. Chung, T.W. Fok Lung, C. Giogha, N. Wang, R. Strugnell, N. Brown, M. Cygler, J. Pearson, E. Hartland. Salmonella effectors SseK1 and SseK3 target death domain proteins in the TNF and TRAIL signaling pathways. Mol. Cell Proteomics (2019) 18(6):1138-1156. *Joint lead author
75. C.M Harding, M.A. Nasr MA, N.E. Scott, G. Goyette-Desjardins, H. Nothaft, A.E. Mayer, S.M. Chavez, J.P Huynh, R.L Kinsella, C.M Szymanski, C.L Stallings, M Segura, M.F Feldman. A platform for glycoengineering a polyvalent pneumococcal bioconjugate vaccine using E. coli as a host. Nat Commun. (2019) Feb 21;10(1):891.
76. J.A Cain, A.L Dale, P. Niewold, W.P Klare, L Man, M.Y White, N.E Scott, S.J Cordwell. Proteomics reveals multiple phenotypes associated with N-linked glycosylation in Campylobacter jejuni. Mol Cell Proteomics. (2019). 18(4):715-734
77. S.L Vogt, R Scholz, Y Peng, R.L Guest, N.E Scott, S.E Woodward, L.J Foster, T.L Raivio, B.B Finlay. Characterization of the Citrobacter rodentium Cpx regulon and its role in host infection. Mol Microbiol. (2019) 111(3):700-716.
78. S. Khurana, M.J. Coffey, A. John, A.D. Uboldi, M.H. Huynh, R.J. Stewart, V.B Carruthers, C. Tonkin, E.D Goddard-Borger, N.E Scott#. Protein O-fucosyltransferase 2–mediated O-glycosylation of the adhesin MIC2 is dispensable for Toxoplasma gondii tachyzoite infection. J Biol Chem (2019) 1;294(5):1541-1553. # corresponding authorship
2018
79. E. Valguarnera, N.E. Scott, P. Azimzadeh, M.Feldman . Surface Exposure and Packing of Lipoproteins into Outer Membrane Vesicles Are Coupled Processes in Bacteroides. mSphere (2018) Nov 7;3(6). pii: e00559-18.
80. C. Kerr, U. Dalwadi, N.E Scott, C. Yip, L.J. Foster, E. Jan. Transmission of Cricket paralysis virus via exosome-like vesicles during infection of Drosophila cells. Scientific reports (2018) Nov 26;8(1):17353
81. N.C. McKenzie, N.E. Scott, A. John, J.M. White, E.D. Goddard-Borger. Synthesis and use of 6,6,6-trifluoro-L-fucose to block core-fucosylation in hybridoma cell lines. Carbohydr Res. (2018) May 23; 465:4-9.
80. C. Kerr, U. Dalwadi, N.E Scott, C. Yip, L.J. Foster, E. Jan. Transmission of Cricket paralysis virus via exosome-like vesicles during infection of Drosophila cells. Scientific reports (2018) Nov 26;8(1):17353
81. N.C. McKenzie, N.E. Scott, A. John, J.M. White, E.D. Goddard-Borger. Synthesis and use of 6,6,6-trifluoro-L-fucose to block core-fucosylation in hybridoma cell lines. Carbohydr Res. (2018) May 23; 465:4-9.
Pre-establishment of Scott lab publications
2017
82. R.H Malty, H Aoki, A Kumar, S Phanse, S Amin, Q Zhang, Z Minic, F Goebels, G Musso, Z Wu, H Abou-Tok, M Meyer, V Deineko, S Kassir, V Sidhu, M Jessulat, N.E Scott, X Xiong, J Vlasblom, B Prasad, L.J Foster, T Alberio, B Garavaglia, H Yu, G.D Bader, K Nakamura, J Parkinson, M Babu. A Map of Human Mitochondrial Protein Interactions Linked to Neurodegeneration Reveals New Mechanisms of Redox Homeostasis and NF-κB Signaling. Cell Syst. (2017) Dec 27;5(6):564-577.e12.
83. N.E Scott and E.L Hartland. Post-translational mechanisms of host subversion by bacterial. Trends in Molecular Medicine (2017). Dec;23(12):1088-1102.
84. R.G. Stacey, M.A Skinnider, N.E Scott, L.J. Foster. A rapid and accurate approach for prediction of interactomes from co-elution data (PrInCE). BMC Bioinformatics. (2017) 18(1):457.
85. S. Lopaticki, A.S.P Yang, A John, N.E Scott, J.P Lingford, M.T O'Neill, S.M Erickson, N.C McKenzie, C Jennison, L.W Whitehead, D.N Douglas, N.M Kneteman, E.D Goddard-Borger, J.A Boddey. Protein O-fucosylation in Plasmodium falciparum ensures efficient infection of mosquito and vertebrate hosts. Nat Commun. (2017) 8(1) :561.
86. N.E Scott#, C Giogha, G.L Pollock, C.L Kenned, A.I Webb, N.A Williamson, J.S Pearson, E.L Hartland. The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J Biol Chem. (2017) 292(42):17337-17350. # corresponding authorship
87. N.E Scott, E.L Hartland. The role of mass spectrometry analysis in bacterial effector characterization. Biochem J. (2017) 474(16):2779-2784
88. C.M Harding, M.R Pulido, G Di Venanzio, R.L Kinsella, A.I Webb, N.E Scott, J Pachón, M.F Feldman. Pathogenic Acinetobacter Species have a Functional Type I Secrection System and Contact-Dependent Inhibition Systems. J Biol Chem. (2017 )292(22):9075-9087.
89. J.M McCoy, R.J Stewart, A.D Uboldi, D Li, J Schröder, N.E Scott, A.T Papenfuss, A.M Lehane, L.J Foster, C.J Tonkin. A Forward-gene c Screen Identifies a Negative Regulator of Rapid Ca2+- dependent Cell Egress in the Intracellular Parasite Toxoplasma gondii. J Biol Chem. (2017) 292(18):7662-7674.
90. N.E. Scott, R.D. Rogers, A. Prudova, N.F. Brown, N. Fortelny, C.M. Overall, and L.J. Foster. Interactome disassembly during apoptosis occurs independent of caspase cleavage. Mol Syst Biol (2017). 13(1):906.
91. K. Creuzburg, C. Giogha, T Wong Fok Lung, N.E. Scott, S. Mühlen, E.L. Hartland, J.S. Pearson. The Type III effector NleD from enteropathogenic E. coli differentiates between host substrates p38 and JNK. Infect Immun. (2017). 85(2): e00620-16.
83. N.E Scott and E.L Hartland. Post-translational mechanisms of host subversion by bacterial. Trends in Molecular Medicine (2017). Dec;23(12):1088-1102.
84. R.G. Stacey, M.A Skinnider, N.E Scott, L.J. Foster. A rapid and accurate approach for prediction of interactomes from co-elution data (PrInCE). BMC Bioinformatics. (2017) 18(1):457.
85. S. Lopaticki, A.S.P Yang, A John, N.E Scott, J.P Lingford, M.T O'Neill, S.M Erickson, N.C McKenzie, C Jennison, L.W Whitehead, D.N Douglas, N.M Kneteman, E.D Goddard-Borger, J.A Boddey. Protein O-fucosylation in Plasmodium falciparum ensures efficient infection of mosquito and vertebrate hosts. Nat Commun. (2017) 8(1) :561.
86. N.E Scott#, C Giogha, G.L Pollock, C.L Kenned, A.I Webb, N.A Williamson, J.S Pearson, E.L Hartland. The bacterial arginine glycosyltransferase effector NleB preferentially modifies Fas-associated death domain protein (FADD). J Biol Chem. (2017) 292(42):17337-17350. # corresponding authorship
87. N.E Scott, E.L Hartland. The role of mass spectrometry analysis in bacterial effector characterization. Biochem J. (2017) 474(16):2779-2784
88. C.M Harding, M.R Pulido, G Di Venanzio, R.L Kinsella, A.I Webb, N.E Scott, J Pachón, M.F Feldman. Pathogenic Acinetobacter Species have a Functional Type I Secrection System and Contact-Dependent Inhibition Systems. J Biol Chem. (2017 )292(22):9075-9087.
89. J.M McCoy, R.J Stewart, A.D Uboldi, D Li, J Schröder, N.E Scott, A.T Papenfuss, A.M Lehane, L.J Foster, C.J Tonkin. A Forward-gene c Screen Identifies a Negative Regulator of Rapid Ca2+- dependent Cell Egress in the Intracellular Parasite Toxoplasma gondii. J Biol Chem. (2017) 292(18):7662-7674.
90. N.E. Scott, R.D. Rogers, A. Prudova, N.F. Brown, N. Fortelny, C.M. Overall, and L.J. Foster. Interactome disassembly during apoptosis occurs independent of caspase cleavage. Mol Syst Biol (2017). 13(1):906.
91. K. Creuzburg, C. Giogha, T Wong Fok Lung, N.E. Scott, S. Mühlen, E.L. Hartland, J.S. Pearson. The Type III effector NleD from enteropathogenic E. coli differentiates between host substrates p38 and JNK. Infect Immun. (2017). 85(2): e00620-16.
2016
92. B.S Weber, S.W. Hennon, M.S. Wright, N.E. Scott, V. de Berardinis, L.J. Foster, J.A. Ayala, M.D. Adams, M.F. Feldman. Genetic Dissection of the Type VI Secretion System in Acinetobacter and Identification of a Novel Peptidoglycan Hydrolase, TagX, Required for Its Biogenesis. MBio. (2016) 7(5): e01253-16.
93. M.J. Bround, R. Wambolt, H. Cen, P. Asghari, R.F. Albu, J. Han, D. McAfee, M. Pourrier, N.E. Scott, L. Bohunek, J.E. Kulpa, S.R. Chen, D. Fedida, R.W. Brownsey, C.H. Borchers, L.J. Foster, T. Mayor, E.D. Moore, M.F. Allard, J.D. Johnson. Cardiac Ryanodine Receptor (Ryr2)-mediated Calcium Signals Specifically Promote Glucose Oxidation via Pyruvate Dehydrogenase. J Biol Chem. (2016) 4;291(45):23490-23505.
93. M.J. Bround, R. Wambolt, H. Cen, P. Asghari, R.F. Albu, J. Han, D. McAfee, M. Pourrier, N.E. Scott, L. Bohunek, J.E. Kulpa, S.R. Chen, D. Fedida, R.W. Brownsey, C.H. Borchers, L.J. Foster, T. Mayor, E.D. Moore, M.F. Allard, J.D. Johnson. Cardiac Ryanodine Receptor (Ryr2)-mediated Calcium Signals Specifically Promote Glucose Oxidation via Pyruvate Dehydrogenase. J Biol Chem. (2016) 4;291(45):23490-23505.
2015
94. W. Elhenawy, N. E. Scott, M.L. Tondo, E.G. Orellano, L.J. Foster, M.F. Feldman. Protein O-linked glycosylation in the plant pathogen Ralstonia solanacearum. Glycobiology. (2015) 26(3):301-11.
95. A. Cameron, S. Huynh, N.E. Scott, E. Frirdich, D. Apel, L.J Foster, C.T. Parker, E.C. Gaynor. High-Frequency Variation of Purine Biosynthesis Genes Is a Mechanism of Success in Campylobacter jejuni. MBio. 2015 6: e00612-15.
96. G. Volkers, L. Worrall, D. Kwan, C. Yu, L. Baumann, E. Lameignere, G. Wasney, N.E. Scott, W. Wakarchuk, L.J. Foster, S. Withers, N. Strynadka. Structure of human ST8SiaIII sialyltransferase provides insight into cell surface polysialylation. Nat Struct Mol Biol. (2015) 22: 627-35.
97. R. Scholz, K. Imami, N.E. Scott, W.S. Trimble, L.J. Foster, B.B Finlay. Novel Host Proteins and Signaling Pathways in Enteropathogenic E. coli Pathogenesis Identified by Global Phosphoproteome Analysis. Mol Cell Proteomics. (2015) 14(7):1927-45.
98. C.M. Harding, M.A. Nasr, R.L. Kinsella, N.E. Scott, J.L Foster, B.S. Weber, S.E Fiester, L.A. Actis, E.N. Tracy, R.S. Munson, M.F. Feldman. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol Microbiol. (2015) 96(5):1023-41.
99. N.E Scott#, L.M Brown, A.R Kristensen, J.L Foster. Development of a computational framework for the analysis of protein correlation profiling and spatial proteomics experiments. J Proteomics. (2015), 118:112-29. # corresponding authorship
95. A. Cameron, S. Huynh, N.E. Scott, E. Frirdich, D. Apel, L.J Foster, C.T. Parker, E.C. Gaynor. High-Frequency Variation of Purine Biosynthesis Genes Is a Mechanism of Success in Campylobacter jejuni. MBio. 2015 6: e00612-15.
96. G. Volkers, L. Worrall, D. Kwan, C. Yu, L. Baumann, E. Lameignere, G. Wasney, N.E. Scott, W. Wakarchuk, L.J. Foster, S. Withers, N. Strynadka. Structure of human ST8SiaIII sialyltransferase provides insight into cell surface polysialylation. Nat Struct Mol Biol. (2015) 22: 627-35.
97. R. Scholz, K. Imami, N.E. Scott, W.S. Trimble, L.J. Foster, B.B Finlay. Novel Host Proteins and Signaling Pathways in Enteropathogenic E. coli Pathogenesis Identified by Global Phosphoproteome Analysis. Mol Cell Proteomics. (2015) 14(7):1927-45.
98. C.M. Harding, M.A. Nasr, R.L. Kinsella, N.E. Scott, J.L Foster, B.S. Weber, S.E Fiester, L.A. Actis, E.N. Tracy, R.S. Munson, M.F. Feldman. Acinetobacter strains carry two functional oligosaccharyltransferases, one devoted exclusively to type IV pilin, and the other one dedicated to O-glycosylation of multiple proteins. Mol Microbiol. (2015) 96(5):1023-41.
99. N.E Scott#, L.M Brown, A.R Kristensen, J.L Foster. Development of a computational framework for the analysis of protein correlation profiling and spatial proteomics experiments. J Proteomics. (2015), 118:112-29. # corresponding authorship
2014
100. R.L Kinsella, N.E Scott, M.F Feldman. Clinical implications of glycoproteomics for Acinetobacter baumannii. Expert Rev Proteomics. (2014), 11: 1-3
101. N.E Scott, N.B Marzook, J.A Cain, N Solis, M Thaysen-Andersen, S.P Djordjevic, N.H Packer, M.R Larsen, S.J Cordwell. Comparative Proteomics and Glycoproteomics Reveal Increased N-Linked Glycosylation and Relaxed Sequon Specificity in Campylobacter jejuni NCTC11168 O. J Proteome Res. (2014), 13(11): 5136-5150.
102. N.E Scott, R.L Kinsella, A.V Edwards, M.R Larsen, S Dutta, J Saba, L.J Foster, M.F Feldman. Diversity within the O-linked protein glycosylation systems of Acinetobacter species. Mol Cell Proteomics. (2014), 13(9): 2354-2370.
103. K.V Lithgow*, N.E Scott*, J.A Iwashkiw, E.L Thomson, L.J Foster, M.F Feldman, J.J Dennis. A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol Microbiol. (2014), 92(1): 116-137 *Joint lead author
101. N.E Scott, N.B Marzook, J.A Cain, N Solis, M Thaysen-Andersen, S.P Djordjevic, N.H Packer, M.R Larsen, S.J Cordwell. Comparative Proteomics and Glycoproteomics Reveal Increased N-Linked Glycosylation and Relaxed Sequon Specificity in Campylobacter jejuni NCTC11168 O. J Proteome Res. (2014), 13(11): 5136-5150.
102. N.E Scott, R.L Kinsella, A.V Edwards, M.R Larsen, S Dutta, J Saba, L.J Foster, M.F Feldman. Diversity within the O-linked protein glycosylation systems of Acinetobacter species. Mol Cell Proteomics. (2014), 13(9): 2354-2370.
103. K.V Lithgow*, N.E Scott*, J.A Iwashkiw, E.L Thomson, L.J Foster, M.F Feldman, J.J Dennis. A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol Microbiol. (2014), 92(1): 116-137 *Joint lead author
2013
104. B.L Parker, M Thaysen-Andersen, N Solis, N.E Scott, M.R Larsen, M.E Graham, N.H Packer, S.J Cordwell. Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J Proteome Res. (2013), 12(12): 5791-5800.
105. N.E Scott, N.J Hare, M.Y White, J Manos, S.J Cordwell. Secretome of transmissible Pseudomonas aeruginosa AES-1R grown in a cystic fibrosis lung-like environment. J Proteome Res. (2013), 12(12): 5357-5369.
106. R.G Lees-Miller, J.A Iwashkiw, N. E Scott, A Seper, E Vinogradov, S Schild, M.F Feldman. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol Microbiol (2013), 89(5): 816-830. Highlighted in the Molecular Microbiology MicroCommentary “Bacteria like sharing their sweets” (2013, 89(5): 811-815).
107. M.A. Musumeci, I Hug, N.E Scott, M.V. Ielmini, L.J. Foster, P.G. Wang, M.F. Feldman. In vitro activity of Neisseria meningitidis PglL O-oligosaccharyltransferase with diverse synthetic lipid donors and a UDP-activated sugar. J. Biol. Chem. (2013). 288(15): 10578-87.
105. N.E Scott, N.J Hare, M.Y White, J Manos, S.J Cordwell. Secretome of transmissible Pseudomonas aeruginosa AES-1R grown in a cystic fibrosis lung-like environment. J Proteome Res. (2013), 12(12): 5357-5369.
106. R.G Lees-Miller, J.A Iwashkiw, N. E Scott, A Seper, E Vinogradov, S Schild, M.F Feldman. A common pathway for O-linked protein-glycosylation and synthesis of capsule in Acinetobacter baumannii. Mol Microbiol (2013), 89(5): 816-830. Highlighted in the Molecular Microbiology MicroCommentary “Bacteria like sharing their sweets” (2013, 89(5): 811-815).
107. M.A. Musumeci, I Hug, N.E Scott, M.V. Ielmini, L.J. Foster, P.G. Wang, M.F. Feldman. In vitro activity of Neisseria meningitidis PglL O-oligosaccharyltransferase with diverse synthetic lipid donors and a UDP-activated sugar. J. Biol. Chem. (2013). 288(15): 10578-87.
2012
108. H. Nothaft, N.E. Scott, E. Vinogradov, X. Liu, R. Hu, B. Beadle, C. Fodor, W.G. Miller, J. Li, S.J. Cordwell, C.M Szymanski. Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol. Cell. Proteomics (2012). 11(11): 1203-19.
109. N.E. Scott, H. Nothaft, A.V.G. Edwards, M. Labbate, S.P. Djordjevic, M.R. Larsen, C.M. Szymanski, S.J. Cordwell. Modification of the Campylobacter jejuni N-linked glycan by EptC-mediated addition of phosphoethanolamine. J. Biol. Chem. (2012). 287(35): 29384-96
110. J.A. Iwashkiw, A. Seper, B.S. Weber, N.E. Scott, E. Vinogradov, C. Stratilo, B. Reiz, S.J. Cordwell, R. Whittal, S. Schild, M.F. Feldman. Identification of a General O-linked Protein Glycosylation System in Acinetobacter baumannii and Its Role in Virulence and Biofilm Formation. PLOS pathogens. (2012) 8(6): e1002758. PMID: 22685409. Highlighted in Nature Review Microbiology research highlight (2012, 10: 523).
109. N.E. Scott, H. Nothaft, A.V.G. Edwards, M. Labbate, S.P. Djordjevic, M.R. Larsen, C.M. Szymanski, S.J. Cordwell. Modification of the Campylobacter jejuni N-linked glycan by EptC-mediated addition of phosphoethanolamine. J. Biol. Chem. (2012). 287(35): 29384-96
110. J.A. Iwashkiw, A. Seper, B.S. Weber, N.E. Scott, E. Vinogradov, C. Stratilo, B. Reiz, S.J. Cordwell, R. Whittal, S. Schild, M.F. Feldman. Identification of a General O-linked Protein Glycosylation System in Acinetobacter baumannii and Its Role in Virulence and Biofilm Formation. PLOS pathogens. (2012) 8(6): e1002758. PMID: 22685409. Highlighted in Nature Review Microbiology research highlight (2012, 10: 523).
2011
111. D.R. Bogema, N.E. Scott, M. Padula, J.L. Tacchi, B.B. Raymond, C. Jenkins, S.J. Cordwell, F.C. Minion, M.J. Walker, S.P. Djordjevic. The sequence TTKF | QE defines the site of proteolytic cleavage in Mhp683, a novel glycosaminoglycan and cilium adhesin of Mycoplasma hyopneumoniae. J. Biol. Chem. (2011) 286(48): 41217-29.
112. N.J. Hare*, N.E. Scott*, E.H.H. Shin, A.M. Connolly, M.R. Larsen, G. Palmisano and S.J. Cordwell. Proteomics of the oxida-tive stress response induced by hydrogen peroxide and paraquat reveals a novel AhpC-like Protein in Pseudomonas aeruginosa. Proteomics (2011) 11(15): 3056-69. *Joint lead author.
113. B.L. Parker, G. Palmisano, A.V. Edwards, M.Y. White, K. Engholm-Keller, A. Lee, N.E. Scott, D, Kolarich, B.D. Hambly, N.H. Packer, M.R. Larsen and S.J. Cordwell. Quantitative N-linked glycoproteomics of myocardial ischemia / reperfusion injury reveals early remodeling in the extracellular environment. Mol. Cell. Proteomics (2011). 10(8): M110.006833.
114. N.E. Scott, B.L. Parker, A.M. Connolly, J. Paulech, A.V. Edwards, B. Crossett, L. Falconer, D. Kolarich, S.P. Djordjevic, P. HØjrup, N.H. Packer, M.R. Larsen and S.J. Cordwell. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, HCD and ETD-MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics (2011) 10(2): M000031-MCP201.
112. N.J. Hare*, N.E. Scott*, E.H.H. Shin, A.M. Connolly, M.R. Larsen, G. Palmisano and S.J. Cordwell. Proteomics of the oxida-tive stress response induced by hydrogen peroxide and paraquat reveals a novel AhpC-like Protein in Pseudomonas aeruginosa. Proteomics (2011) 11(15): 3056-69. *Joint lead author.
113. B.L. Parker, G. Palmisano, A.V. Edwards, M.Y. White, K. Engholm-Keller, A. Lee, N.E. Scott, D, Kolarich, B.D. Hambly, N.H. Packer, M.R. Larsen and S.J. Cordwell. Quantitative N-linked glycoproteomics of myocardial ischemia / reperfusion injury reveals early remodeling in the extracellular environment. Mol. Cell. Proteomics (2011). 10(8): M110.006833.
114. N.E. Scott, B.L. Parker, A.M. Connolly, J. Paulech, A.V. Edwards, B. Crossett, L. Falconer, D. Kolarich, S.P. Djordjevic, P. HØjrup, N.H. Packer, M.R. Larsen and S.J. Cordwell. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, HCD and ETD-MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol. Cell. Proteomics (2011) 10(2): M000031-MCP201.
2010
115. N.E. Scott, A. Deutscher, L. Falconer, S.P. Djordjevic and S.J. Cordwell. Mass spectrometric characterization of the Campylobacter jejuni adherence factor CadF reveals post-translational processing that removes immunogenicity while retaining fibronectin binding. Proteomics (2010) 10(2): 277-288.
2009
116. N.E. Scott, D. Bogema, A. Connolly, L. Falconer, S.P. Djordjevic and S.J. Cordwell. Mass spectrometric characterization of the surface-associated 42 kDa lipoprotein JlpA as a glycosylated antigen in strains of Campylobacter jejuni. J Proteome Res. (2009) 8(10): 4654-4664.
117. N.E. Scott and S.J. Cordwell. Campylobacter proteomics: guidelines, challenges and future perspectives. Expert Revs. Prote-omics (2009) 6(1): 61-74.
117. N.E. Scott and S.J. Cordwell. Campylobacter proteomics: guidelines, challenges and future perspectives. Expert Revs. Prote-omics (2009) 6(1): 61-74.
2008
118. S.J. Cordwell, A.C.L. Len, R.G. Touma, N.E. Scott, L. Falconer, D. Jones, A. Connolly, B. Crossett and S.P. Djordjevic. Identi-fication of surface-associated proteins from Campylobacter jejuni using complementary proteomics technologies. Proteomics (2008) 8(1): 122-39.